Sharp regression discontinuity with scikit-learn models#
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.gaussian_process.kernels import ExpSineSquared, WhiteKernel
from sklearn.linear_model import LinearRegression
import causalpy as cp
%config InlineBackend.figure_format = 'retina'
Load data#
data = cp.load_data("rd")
data.head()
| x | y | treated | |
|---|---|---|---|
| 0 | -0.932739 | -0.091919 | False |
| 1 | -0.930778 | -0.382663 | False |
| 2 | -0.929110 | -0.181786 | False |
| 3 | -0.907419 | -0.288245 | False |
| 4 | -0.882469 | -0.420811 | False |
Linear, main-effects model#
result = cp.RegressionDiscontinuity(
data,
formula="y ~ 1 + x + treated",
model=LinearRegression(),
treatment_threshold=0.5,
)
fig, ax = result.plot()
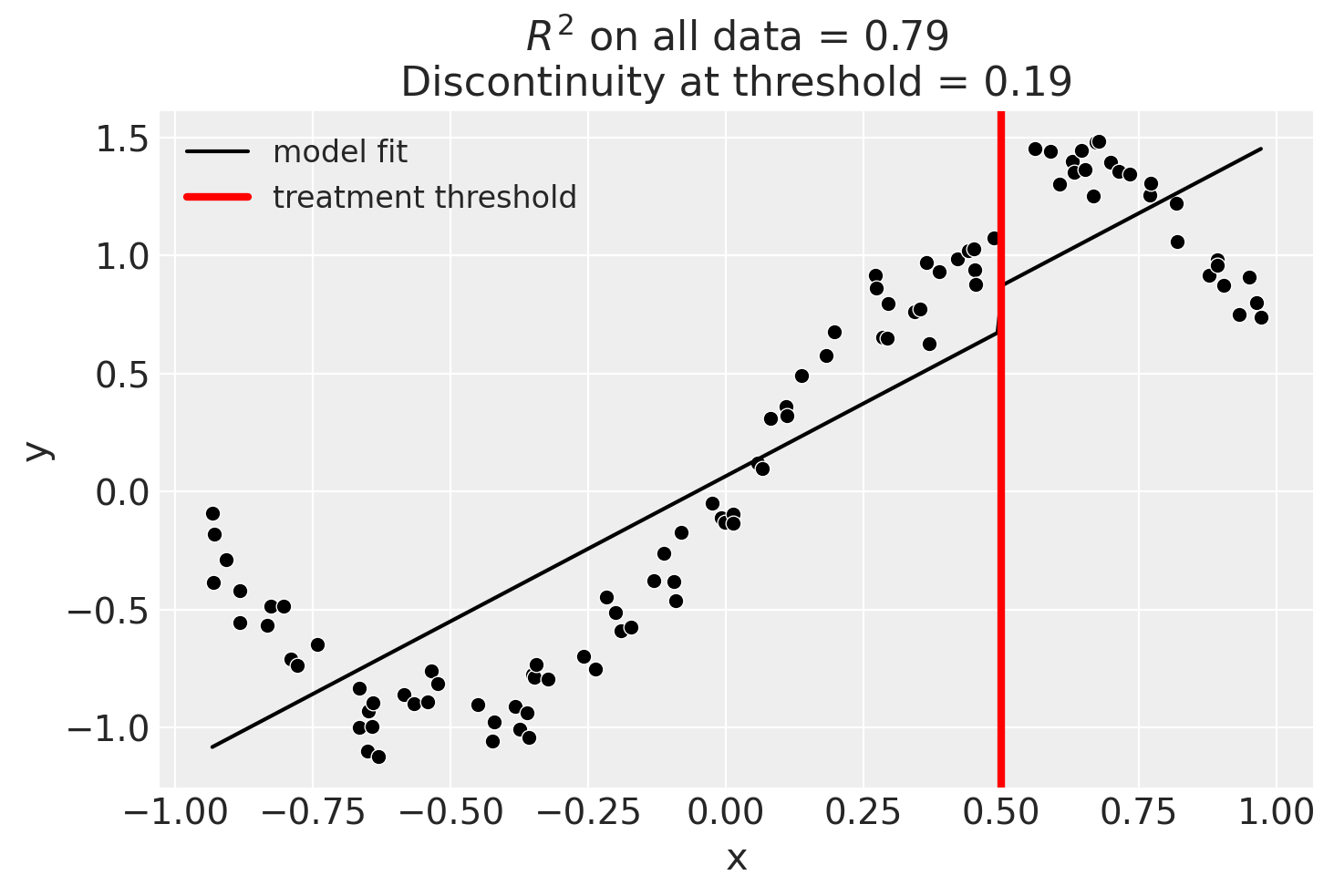
result.summary(round_to=3)
Difference in Differences experiment
Formula: y ~ 1 + x + treated
Running variable: x
Threshold on running variable: 0.5
Results:
Discontinuity at threshold = 0.19
Model coefficients:
Intercept 0
treated[T.True] 0.19
x 1.23
Linear, main-effects, and interaction model#
result = cp.RegressionDiscontinuity(
data,
formula="y ~ 1 + x + treated + x:treated",
model=LinearRegression(),
treatment_threshold=0.5,
)
result.plot();
Though we can see that this does not give a good fit of the data almost certainly overestimates the discontinuity at threshold.
result.summary(round_to=3)
Difference in Differences experiment
Formula: y ~ 1 + x + treated + x:treated
Running variable: x
Threshold on running variable: 0.5
Results:
Discontinuity at threshold = 0.92
Model coefficients:
Intercept 0
treated[T.True] 2.47
x 1.32
x:treated[T.True] -3.11
Using a bandwidth#
One way how we could deal with this is to use the bandwidth kwarg. This will only fit the model to data within a certain bandwidth of the threshold. If \(x\) is the running variable, then the model will only be fitted to data where \(threshold - bandwidth \le x \le threshold + bandwidth\).
result = cp.RegressionDiscontinuity(
data,
formula="y ~ 1 + x + treated + x:treated",
model=LinearRegression(),
treatment_threshold=0.5,
bandwidth=0.3,
)
result.plot();
We could even go crazy and just fit intercepts for the data close to the threshold. But clearly this will involve more estimation error as we are using less data.
result = cp.RegressionDiscontinuity(
data,
formula="y ~ 1 + treated",
model=LinearRegression(),
treatment_threshold=0.5,
bandwidth=0.3,
)
result.plot();
Using Gaussian Processes#
Now we will demonstrate how to use a scikit-learn model.
kernel = 1.0 * ExpSineSquared(1.0, 5.0) + WhiteKernel(1e-1)
result = cp.RegressionDiscontinuity(
data,
formula="y ~ 1 + x + treated",
model=GaussianProcessRegressor(kernel=kernel),
treatment_threshold=0.5,
)
fig, ax = result.plot()
Using basis splines#
result = cp.RegressionDiscontinuity(
data,
formula="y ~ 1 + bs(x, df=6) + treated",
model=LinearRegression(),
treatment_threshold=0.5,
)
fig, ax = result.plot()
result.summary(round_to=3)
Difference in Differences experiment
Formula: y ~ 1 + bs(x, df=6) + treated
Running variable: x
Threshold on running variable: 0.5
Results:
Discontinuity at threshold = 0.41
Model coefficients:
Intercept 0
treated[T.True] 0.407
bs(x, df=6)[0] -0.594
bs(x, df=6)[1] -1.07
bs(x, df=6)[2] 0.278
bs(x, df=6)[3] 1.65
bs(x, df=6)[4] 1.03
bs(x, df=6)[5] 0.567
Effect Summary Reporting#
For decision-making, you often need a concise summary of the causal effect. The effect_summary() method provides a decision-ready report with key statistics. Note that for Regression Discontinuity, the effect is a single scalar (the discontinuity at the threshold), similar to Difference-in-Differences.
Note
OLS vs PyMC Models: When using OLS models (scikit-learn), the effect_summary() provides confidence intervals and p-values (frequentist inference), rather than the posterior distributions, HDI intervals, and tail probabilities provided by PyMC models (Bayesian inference). OLS tables include: mean, CI_lower, CI_upper, and p_value, but do not include median, tail probabilities (P(effect>0)), or ROPE probabilities.
# Generate effect summary for the final model (basis splines)
stats = result.effect_summary()
stats.table
| mean | ci_lower | ci_upper | p_value | |
|---|---|---|---|---|
| discontinuity | 0.407683 | 0.388323 | 0.427043 | 0.0 |
# View the prose summary
print(stats.text)
The discontinuity at threshold was 0.41 (95% CI [0.39, 0.43]), with a p-value of 0.000.
# You can specify the direction of interest (e.g., testing for an increase)
stats_increase = result.effect_summary(direction="increase")
stats_increase.table
| mean | ci_lower | ci_upper | p_value | |
|---|---|---|---|---|
| discontinuity | 0.407683 | 0.388323 | 0.427043 | 0.0 |
