Difference in Differences with pymc models#
Note
This example is in-progress! Further elaboration and explanation will follow soon.
import arviz as az
import causalpy as cp
%load_ext autoreload
%autoreload 2
%config InlineBackend.figure_format = 'retina'
seed = 42
Load data#
df = cp.load_data("did")
df.head()
| group | t | unit | post_treatment | y | |
|---|---|---|---|---|---|
| 0 | 0 | 0.0 | 0 | False | 0.897122 |
| 1 | 0 | 1.0 | 0 | True | 1.961214 |
| 2 | 1 | 0.0 | 1 | False | 1.233525 |
| 3 | 1 | 1.0 | 1 | True | 2.752794 |
| 4 | 0 | 0.0 | 2 | False | 1.149207 |
Run the analysis#
Note
The random_seed keyword argument for the PyMC sampler is not necessary. We use it here so that the results are reproducible.
result = cp.DifferenceInDifferences(
df,
formula="y ~ 1 + group*post_treatment",
time_variable_name="t",
group_variable_name="group",
model=cp.pymc_models.LinearRegression(sample_kwargs={"random_seed": seed}),
)
Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (4 chains in 4 jobs)
NUTS: [beta, y_hat_sigma]
Sampling 4 chains for 1_000 tune and 1_000 draw iterations (4_000 + 4_000 draws total) took 1 seconds.
Sampling: [beta, y_hat, y_hat_sigma]
Sampling: [y_hat]
Sampling: [y_hat]
Sampling: [y_hat]
Sampling: [y_hat]
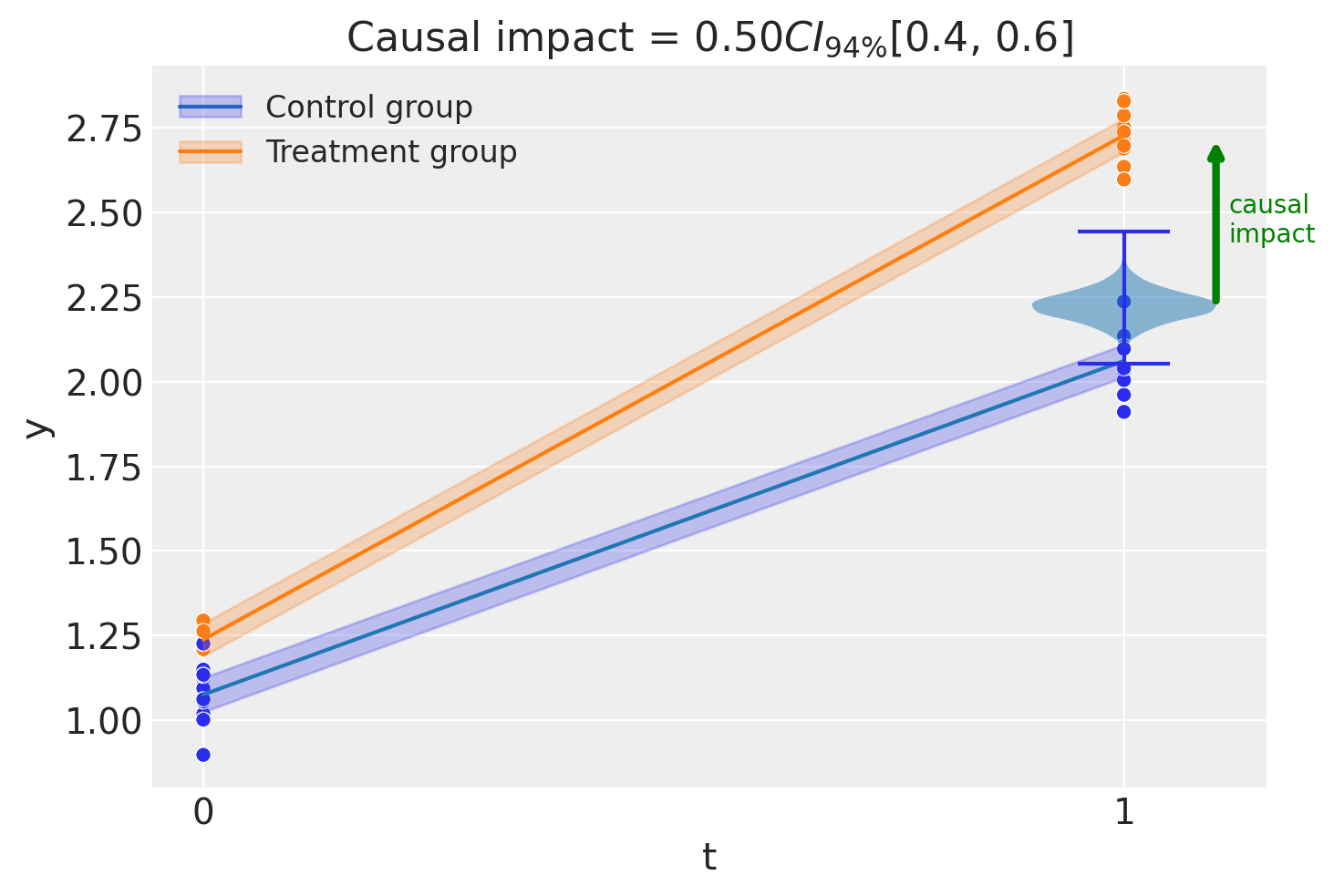
fig, ax = result.plot()
result.summary()
===========================Difference in Differences============================
Formula: y ~ 1 + group*post_treatment
Results:
Causal impact = 0.50$CI_{94\%}$[0.4, 0.6]
Model coefficients:
Intercept 1.1, 94% HDI [1, 1.1]
post_treatment[T.True] 0.99, 94% HDI [0.92, 1.1]
group 0.16, 94% HDI [0.094, 0.23]
group:post_treatment[T.True] 0.5, 94% HDI [0.4, 0.6]
y_hat_sigma 0.082, 94% HDI [0.066, 0.1]
ax = az.plot_posterior(result.causal_impact, ref_val=0)
ax.set(title="Posterior estimate of causal impact");
Effect Summary Reporting#
For decision-making, you often need a concise summary of the causal effect. The effect_summary() method provides a decision-ready report with key statistics. Note that for Difference-in-Differences, the effect is a single scalar (average treatment effect), unlike time-series experiments where effects vary over time.
# Generate effect summary
stats = result.effect_summary()
stats.table
| mean | median | hdi_lower | hdi_upper | p_gt_0 | |
|---|---|---|---|---|---|
| treatment_effect | 0.503779 | 0.504367 | 0.401661 | 0.607651 | 1.0 |
print(stats.text)
The average treatment effect was 0.50 (95% HDI [0.40, 0.61]), with a posterior probability of an increase of 1.000.
You can customize the summary with different directions and ROPE thresholds:
Direction: Test for increase, decrease, or two-sided effect
Alpha: Set the HDI confidence level (default 95%)
ROPE: Specify a minimal effect size threshold
# Example: Two-sided test with ROPE
stats = result.effect_summary(
direction="two-sided",
alpha=0.05,
min_effect=0.3, # Region of Practical Equivalence
)
stats.table
| mean | median | hdi_lower | hdi_upper | p_two_sided | prob_of_effect | p_rope | |
|---|---|---|---|---|---|---|---|
| treatment_effect | 0.503779 | 0.504367 | 0.401661 | 0.607651 | 0.0 | 1.0 | 0.99925 |
print("\n" + stats.text)
The average treatment effect was 0.50 (95% HDI [0.40, 0.61]), with a posterior probability of an effect of 1.000.
